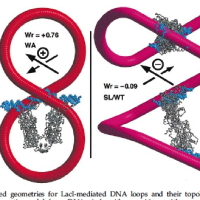
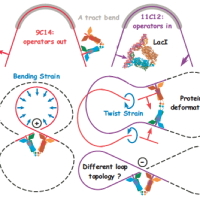
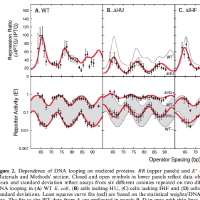
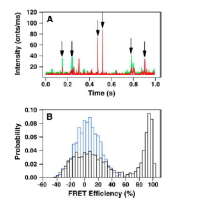
Current Teaching with representative syllabi:
Chemistry 277, Bioanalytical Chemistry Laborator: Syllabus. The course covers fundamental analytical techniques like absorbance, fluorescence, titrations, and chemical kinetics. We also make nanoparticles, and we are experimenting in silico with the Einstein solid.
Chemistry 271, General Chemistry and Energetics: Syllabus.This is a course
covering the traditional material for the second half of general chemistry,
but given in the fourth semester of the undergraduate series, after organic
chemistry instead of before. I also include special topics applications like DNA hybridization
thermodynamics and the redox reactions driving anaerobic metabolism.
BSCI 338F is the seminar class associated with the iGEM team, vide supra.
We have converted our graduate Nucleic Acids class to two 2-credit half-semester modules, Biochemistry 661 and 662, sometimes tag-teamed with
Paul Paukstelis,
Dorothy Beckett, or
Kwaku Dayie.
At various times I have taught Biochem 461 (Biomolecules and Enzymes), 463 (Biochemistry and Physiology), 465 (Biological Information Processing), and parts of 462 (Metabolism).
Here is a TEDxUMD talk (15 min) from Spring, 2014 on my teaching philosophy.
8800+ views, although other faculty may sympathize with the fact that I have not walked it all the way through yet.