Teaching Resources in Biochemistry:
Jason Kahn, University of Maryland College Park.
Disclaimer: I know this area could be better organized. I know some of the content is out of date. It will gradually improve. Let me know if specific things that you need are broken.
Nucleic Acid Sequence, Stability, Structure, Dynamics,
Enzymology
- RNA/DNA structure
- The Watson-Crick base pairs. Interchangeability within a helix, (pseudo)symmetry
axes.
- Hand-drawn sketches of the geometric
origin of helicity and DNA microheterogeneity/bending.
- How a helix grows from a base pair.
- Jmol visualization of B-DNA and A-RNA.
- Why there's no B-form RNA.
- Jmol visualization of tRNA.
- Structure of a psoralen crosslink.
- Rasmol on Tertiary structure in tRNA.
- Sketch of RNA secondary and tertiary structure.
- Here are web sites for RNA structure, function, folding:
- DNA Flexibility and Bending
- Sketch of DNA bending.
- Sketch of DNA flexibibility and cyclization.
- Wormlike coil model
for DNA.
- Informal notes on DNA topology.
- Sketch of the definition of linking number.
- Twist-writhe interconversion in DNA supercoiling.
- Sketch of the shapes assumed by supercoiled DNA.
- Nucleosomes and Chromatin
- Basic nucleosome structure.
- 30 nm fiber structure
- Tetrasome structure (model for 30 nm fiber)
- Histone code proposal
- Patterns of chromatin modification
- Bioinformatics
- Intro to BLAST (from Chuck Delwiche).
- Web site for tutorials on bioinformatics.
- Mathematics and Simulations of Hybridization, protein-DNA binding, kinetic partitioning.
- Page of math on simple binding curves. And here is a simple
spreadsheet illustrating the simple Ybar = P/(P+Kd) binding isotherm.
- Spreadsheet and mathematics for two-state hybridization
and melting curve predictions.
- Detailed discussion of proofreading through
kinetic partitioning.
- Processes
- DNA polymerase structures, mechanism, active sites.
- T7 DNA polymerase structure and fingers closing.
- Organization of the Pol III holoenzyme complex.
- PDF of images of the trombone model, DnaA structure, and the initiation
of replication.
- (External, high tech.) Animations
of the pol III replisome.
- RNA Polymerase cartoons and structures.
- Notes on RNAi.
- Sketch of replication-mediated homologous recombination
in E. coli.
- Sketch of recombination-mediated repair of collapsed replication forks
in E. coli.
- Movies of the first
step of translation from molecular dynamics. And the more lighthearted,
but still informative, 1971 dance
version of the whole process of translation.The movie was sponsored by the late Kent Wilson, of UCSD.
Tools and Tutorials for Molecular Structure:
This file links to a variety of small tutorials/examples focusing on visualization
of biomolecules in three dimensions, using the program RasMol.
A very brief note on philosophy
These pages are intended to help the user learn to use RasMol as a research tool, not just to
look at pictures of molecules. For this reason, I have decided to provide lists of commands
rather than Rasmol scripts (these are very different) and to use RasMol rather than Chime,
so that the user has access to the process (indeed, cannot avoid the process) by which these
pictures were all created. Once methods for viewing and integrating Rasmol scripts into Chime
presentations become more robust, maybe I will switch over. The teach a person to fish paradigm.
Note: perhaps I flatter myself, but if you link to this page or otherwise
make substantive use of the content, please let me know and/or link/give credit
appropriately. Suggestions are always welcome. The creation of this content was part of the educational
part of an NSF Career Award Proposal.
Rasmol/Chime/Jmol installation and use
Update 2005: The free Jmol viewer appears to be a viable replacement for most
of Rasmol and especially Chime.
To view structures in PDB (= Protein Data Bank) format you need a program which
understands PDB. The free options in most common use are RasMol and Chime. Rasmol
is a stand-alone program available for Mac, Windows, and Unix platforms. Chime
is a plug-in which as of this writing works only with recent versions of Netscape.
The Protein Explorer
is essentially an environment used to run Chime more intuitively, with a lot of
typical commands pre-loaded. It makes extensive use of javascripts for communication
between Chime and the user.
To view structures on/from the Web, you have to configure your browser to
launch a PDB viewer program. To do this, you have several options. This server
(www.biochem.umd.edu) supports the experimental mime type chemical/x-pdb. If
you configure your browser correctly (see below), you can bring up a pdb viewer
(either Rasmol or Chime) directly upon clicking on a PDB file link. Otherwise,
you will be asked to save the file and can then read it in to a viewer (Rasmol)
from outside your browser. On some platforms (e.g. Silicon Graphics, probably
other X-windows), you need to tell Netwscape to open a shell window which then
in turn runs Rasmol, otherwise the command window will not appear (Netscape
intercepts the application's desire to open a window). One option that should
always work is to launch Rasmol on its own, save the pdb file as text (download
by holding the mouse down on the link (Mac) or using the alt key (SGI)), and
then open the pdb file from within Rasmol.
There is an excellent resource on Rasmol and Chime, including pointers to
download locations and many examples, at
U Mass. This site showcases much more sophisticated presentations than are
provided here, but at some cost to simplicity and interactivity. The Protein
Explorer is emphasized at the site.
You can get the Chime plug-in to display structures from
within Netscape from
MDLI .
IMHO (= in my humble opinion, for "newbies"), Rasmol is better, especially
if you have a pre-1994 computer. Nothing
can crash a computer like Netscape, esp. w/ plug-ins, and Chime doesn't
seem to provide a user-accessible command line, although it can read more
file formats and is apparently better for advanced scripting within Web pages. Rasmol can be run on its
own after you have quit Netscape or whatever and re-connected your phone line,
and the command line gives you more control and immediate feedback.
You can even just ftp the pdb file and run Rasmol without running a browser
at all, but any computer that can't run Netscape will probably choke on
any complicated Rasmol work.
For more rasmol, other PDB viewers, and a great deal more, see
The NIH Guide to Molecular Modeling . It's difficult navigating but
there's a wealth of information there.
Configuration directions for using Rasmol and Chime:
- If it isn't already available,
get Rasmol (it's freely downloadable) and install/compile it on your
computer.
- For Mac/SGI Netscape, set helper applications or plug-in choices under
"Options/General/Helpers", such that the chemical/x-pdb file type
or any file that ends in ".pdb" is opened by Rasmol. To enable the use
of the command window under X-windows, use the line below as the
"helper application" (thank you very much to Roger Sayle for this info):
xterm -e rasmol %s
For Windoze or Internet
Explorer, Microsoft probably lets you do something similar but you're on your
own as to exactly how.
If you install the Chime plug-in, you choose to launch the plug-in essentially as above. You can go back and forth if you wish -- I have both installed but
usually use RasMol.
Some UNIX setups may require you to change your .mailcap file. Include the line:
chemical/x-pdb ; rasmol %s
If you install Chime, it will modify the .mailcap file or otherwise take
over.
For Jmol: Download, install as plugin (more later if necessry).
Biochem 461/463-oriented materials:
If you are using Netscape/Rasmol as above, download the pdb file first and
thereby launch Rasmol. Then go to the appropriate command file and cut and
paste line by line or larger
chunks from Netscape to the command
line in Rasmol (you may be able to choose "Paste" even when the structure
window is uppermost).
Here is a rather arcane spreadsheet rationalizing why irreversible steps in
a pathway are the right places to control flux.
Structures of amino acids and proteins
Loren Williams' teaching pages at Georgia Tech have a wealth of material in a vein similar to
what you will find here.
Protein conformational analysis: the Ramachandran plot
The Ramachandran plot is used to illustrate the conformational space
accessible to proteins, the energetics of different choices of phi/psi
angles. This page contains an illustrated version
with links to pdb files. Note that this is an example of the fact that just
because you have a pdb file doesn't mean the structure shown can actually
form.
- The Graphical table, with phi/psi angles and links. Note this is a very large page (~1 MB) and will take a while to load.
Here is a short explanation of why proline breaks alpha helices, which
is not due to its restricted accessible area on the
Ramachandran plot.
Alpha helix and beta sheet structures
The files below are provided as examples of idealized secondary
structures.
NEW: These little tutorials have been recoeed for jmol but may not have been fully debugged.
Here is the alpha helix.
Here is the same alpha helix in reality in the GCN4-DNA co-crystal.
Here is the beta sheet.
Here is the beta sheet in the protein tendamistat .
The .txt files below are lists of Rasmol commands. On a Mac or SGI, you can cut-and-paste
commands
into the command window as I do in class -- take chunks delimited by # signs or pause statements,
or go line-by-line. Pasting also works in newer versions of Jmol, and it uses essentially the same command language. On Windows, pasting doesn't work and you will have to save the file
as a script and then "script file_name" to run it. You will need to give the complete path.
The .scr file is a Rasmol
script which recreates a particular view. The .pdb files are the actual molecular data files, essentially
xyz coordinates of the atoms. These are just text files -- go ahead and look.
Protein supersecondary and tertiary structure example: TIM et al.
"TIM" is triose phosphate isomerase, a key enzyme in glycolysis. Our
interest is in TIM as a prototype alpha/beta barrel protein and an
example of protein tertiary structure.
- The PDB file (2ypi.pdb) for TIM, with substrate analog
2-phosphoglycolate. Or click here to download as text.
- The Rasmol commands for displaying one of
the two molecules in the PDB file.
- A gif file showing approximately what you should get after running the script
commands.
Myohemerythrin is an oxygen-binding protein, of interest to us an an
example of a four-helix bundle.
- The PDB file (2mhr.pdb) for
myohemerythrin.
- The Rasmol commands for displaying
a simple secondary structure representation. The second part of the
script lets you "slab" through the molecule to see how crowded the
apparently rather open core really is.
- A gif file showing approximately what you should get after running the script
commands.
The transthyretin dimer (the protein formerly known as prealbumin), has two beta sheets
packed against each other.
- The PDB file (1tta.pdb) for
transthyretin.
- The Rasmol commands for displaying
a simple secondary structure representation, using the same color coding as for
myohemerythrin.
- A gif file showing approximately what you should get after running the script
commands.
Hexokinase is a famous enzyme which clamps down on glucose in order to
phosphorylate it in the first step of glycolysis. Mathews and van Holde
show the bottom half of
this protein (Fig. 6.16c) as an example of mixed alpha helix/beta sheet structure.
Note that Rasmol's (and Insight's) structure analysis algorithms don't find quite
as much beta sheet as the text.
- The PDB file (2yhx.pdb) for
the "open" form of hexokinase, with a bound inhibitor of glucose binding (OTG).
The coordinates of the OTG were used to model the position of glucose below.
- The PDB file (1hkg.pdb) for
the "closed" form of hexokinase. Note glucose was not visible in the
original X-ray structure, and the secondary structure is not as refined.
- The Rasmol commands for displaying
a simple secondary structure representation, using the same color coding as for
myohemerythrin.
- Images showing approximately what you should
get after running the script
commands for the closed and open form.
Quaternary structure and allostery
Hemoglobin is the most studied protein in the world. Biochemical,
biophysical, and crystallographic studies have illuminated many of the
tertiary and quaternary changes accompanying cooperative oxygen binding.
- The deoxy form.
Note the relatively large central cavity, and domed hemes, as well as
the relative positions of the end of the beat subunit FG corner (His97)
and the alpha subunit CD corner (Thr41-Pro44).
- The oxy form, modified from the
original pdb to show all four subunits.
Note the relatively small central cavity, and flattened hemes, as well as
the shift of the end of the beta subunit FG corner (His97), which now approaches
alpha Thr38. This rearrangement (and the consequent breaking of salt bridges) is part
of an overall rotation of one alpha-beta dimer with respect to the
other.
- This file of Rasmol commands should work for
both forms.
- The side-by-side comparison shows that local (heme) changes and quaternary
changes are much easier to see than the rather subtle tertiary changes
which mediate the cooperativity. It may help to look at this first and
then go to the rasmol files.
- Here is a spreadsheet allowing you to graph fractional saturation as a function
of binding constants, R and T preferences, BPG, and so forth. Equations taken
from van Holde, "Physical Biochemistry". (Download to disk using right-mouse
or control-mouse, then open in Excel.)
PDB files of nucleic acids and protein-nucleic acid
complexes
I've made available
a large collection of
PDB files,
mainly of RNA, DNA, RNA/DNA binding proteins,
and protein-RNA/DNA complexes. These were
downloaded from the Brookhaven Protein Data
Bank Web site some time ago, so there may have been some evolution of the files after they moved to the new
PDB.
Biochemistry 465, 661/662, and old 674:
- Follow this link to the A and B forms of DNA
and RNA tutorial directory.
- Follow this link to the protein-DNA interaction
tutorial directory.
- Follow this link to the protein-RNA interaction tutorial
directory.
- Here is a PDB
file of tRNA.
Try using Rasmol or your own favorite program to do coloring by group (to
emphasize which helices stack on each other), viewing cartoons and sticks,
and spacefilling residues 22 and (Boolean or) 46 and (Boolean or) 13, which
form a triple base pair that holds the molecule together.
Here is a result from the open-source, cross-platform program PyMol.
Alternatively, here is a Jmol page that shows you most of the same things.
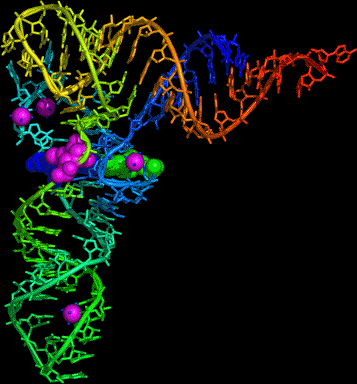
- The structure of the nucleosome.
-
Loren Williams' teaching pages at Georgia Tech have a wealth of material
in a vein similar to what you will find here. Coverage is especially strong
for DNA-drug and DNA-protein interaction.
- The Protein Data Bank's Molecule
of the Month
is often worthwhile.
Here is their view of the nucleosome.
Up to J. Kahn's Home Page

