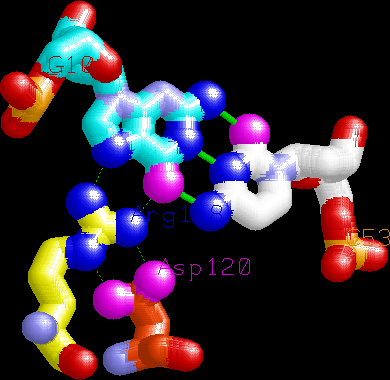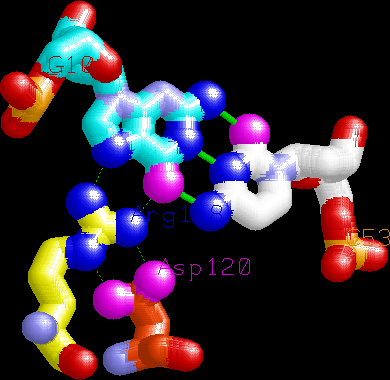Examples of Protein-DNA Co-Complexes
This page is a skeletal tutorial on the structures three classes of
DNA binding proteins and their interactions with DNA, using the Rasmol program to display
three-dimensional structures (here, "PDB files", where PDB = protein data bank format).
Rasmol stuff created by Jason Kahn and Hannah Chen from structures and papers from the
Pabo, Harrison, and Burley groups.
Key concepts
- Understand the general structural features of protein-DNA complexes
and see how the protein recognizes a
DNA molecule in a sequence-specific manner.
- Be able to recognize the three classes of DNA binding motifs illustrated
here, which are the helix-turn-helix, the zinc finger, and the leucine
zipper.
Contents
- This page: directions and sample views (these can be enjoyed without running
the viewer program, but you could look at a book instead).
- HTH-DNA PDB file, 1lmb.pdb,
the structure of the lambda repressor interacting with DNA by Beamer and
Pabo,
along with a file of rasmol commands
for display of important features. And here is essentially the same
content adapted to jmol.
- Zinc finger-DNA PDB file,
1aay.pdb, the structure of the Zif268-DNA complex first crystallized by
Pavletich
and Pabo and subsequently refined further by Pabo's group, with accompanying
command file. Here is a Pymol
session (.pse file) that showcases some of the important features.
And in jmol (under construction).
- bZIP protein-DNA PDB file,
the GCN4 protein by Ellenberger, Brandl, Struhl, and Harrison, with accompanying
command file. And here it
is in Jmol.
- human TBP-DNA PDB file,
from the structure by Nikolov et al. (Burley lab), with accompanying command
file. This structure is very similar to yeast and arabadopsis TBP-DNA
complexes, including those in multi-protein complexes, solved by several
groups. Here is a pymol session file and a movie.
- And here is a link to a separate page on the nucleosome.
- When I create tutorials like this, I work them out by testing commands in the Jmol console (control click in
the window to bring up a menu, ten choose Console). Two good sources for syntax are
at Sourceforge
and
St. Olaf's.
Up to teaching resources
page.
- If you haven't worked out how to use Rasmol, see the Installation Page.
- Load the molecule of your choice into RasMol (either automatically within
Netscape or by hand). The command list includes a line on how to do this but
doesn't do it automatically (because of path issues on the Mac).
- Open the the command list. It might be best to open it as a separate
window, which should happen automagically (if not, on the Mac, hold the mouse button down until the menu pops up).
- Copy and paste from command list file to Rasmol window, or download the file and
do it from a text editor (any word processor will do). On Windows,
pasting doesn't work and you need to save parts of the file and run them
as scripts (use the Notepad). You may need to give a full path for the
script name.
Results
|
Here are gif images generated from the PDB files and scripts above.
The images in the left column emphasize the overall structures of
the three complexes. The right column shows zoomed-in images of
some details of the protein-DNA interactions used in sequence-specific
recognition by the three proteins.
|
|---|
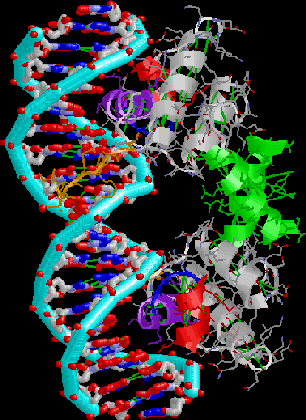
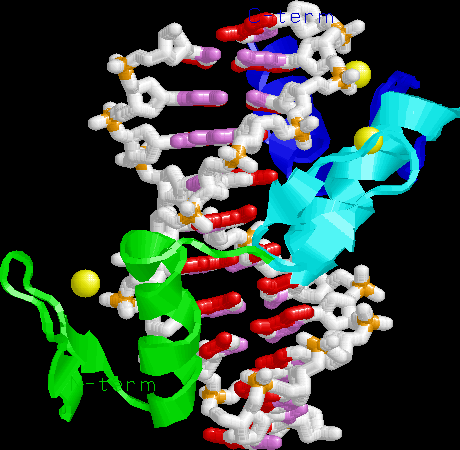
Lambda repressor. Note HTH motif, N-terminal arm, symmetric dimer binding
successive major grooves of a nearly-symmetric DNA site.
|
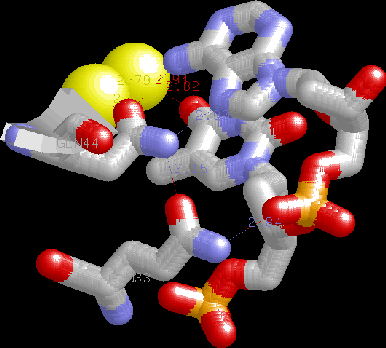
HTH detail. Note the two glutamines H-bonding to each other, specific H-bonds to A,
water-mediated bridge
to adjacent T, H-bonding of packing helix Gln to phosphate backbone
helping to cement recognition helix into the major groove.
|
Zinc finger-DNA. Note the three alpha helices pointing into the major
groove, recognizing 3 bp each. The Zn binding domain is a structural
element for protein folding, not directly involved in DNA binding.
|
Example of a canonical Arginine-Guanine interaction made by the tip
of the first finger. Note the buttressing
aspartate, which helps position the guanidinium group.
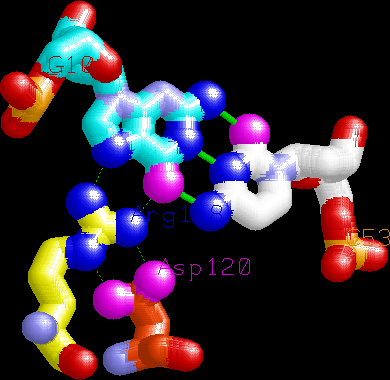
|
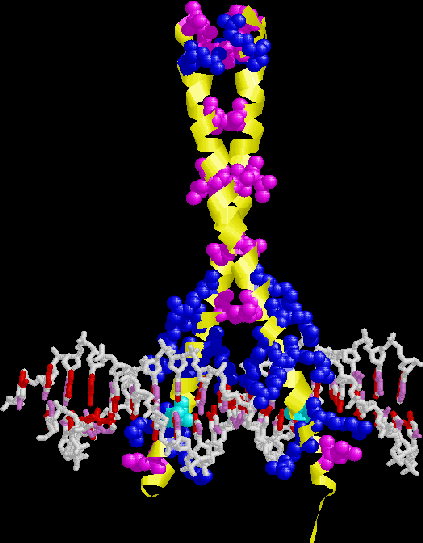
GCN4. Note leucine zipper, basic region, recognition
of adjacent major grooves with "chopstick" motif.
|
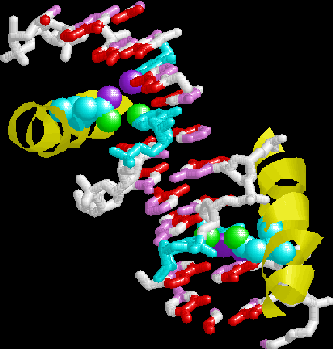
GCN4 detail. Note the two symmetry-related asparagines
recognizing major groove base pair edges. The script
also illustrates an Arg-Gua interaction.
|
TBP. Note the dramatic DNA bending. The DNA projects toward you
on the left, back on the right.
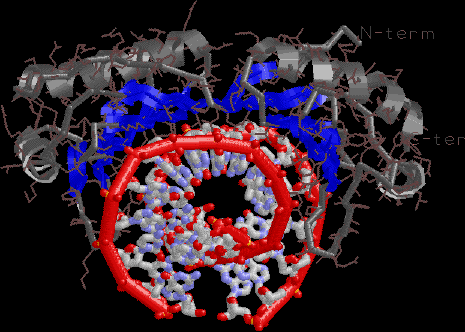
|
TBP view emphasizing the hydrophobic, relatively flat beta sheet
recognition surface.
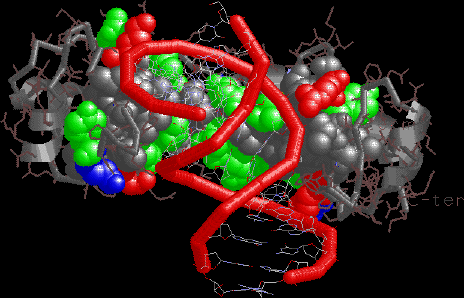
|
TBP view emphasizing the minor groove interaction surface
and dramatic unwinding.
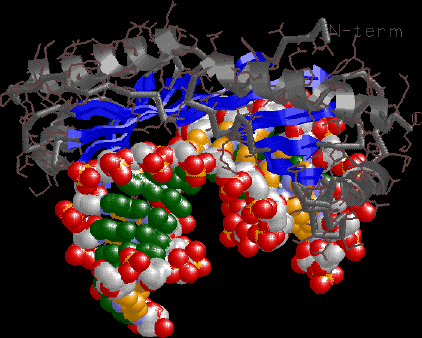
|
TBP detail showing one of the Phe intercalation sites on the
"stirrups" bending the DNA into the major groove.
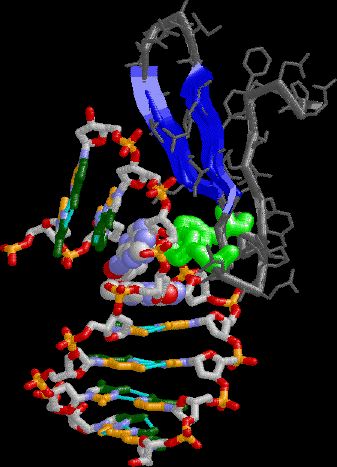
|
Last modified 10/15/97, Jason D. Kahn