Proposal for RNAP Transcription Complex Stability
Numerous X-ray diffraction studies have attempted
to depict accurate views of a transcribing DNA dependent- RNA polymerase.
Watson-Crick interactions occur between transient DNA-RNA hybrids constructed
from template-dependent RNA synthesis (Severinov 2001). Despite this,
uncertainty remains about the required molecular components involved in
the processivity and stability of the RNA polymerase transcription complex
(Severinov 2001). Evidence suggests that the unphosphorylated RNAP
IIa subunit of Rbp 1 initially binds to a promoter, and the phosphorylated
RNAP IIo subunit carries out elongation (Weaver 2002). During formation
of the promoter-complex, RNAPs produce short, abortive nascent RNA oligomers
ranging from 2-8 nucleotides long (Severinov 2001). When an RNA oligomer
of 9 bases is created, it is stably associated with the polymerase transcription
complex, and the RNAP clears the promoter region to processively elongate
the chain (Severinov 2001). Once RNAP encounters an RNA hairpin followed
by a run of uridines, the polymerase pauses and releases nucleic acids because
the stability of the transcription complex is altered (Severinov 2001).
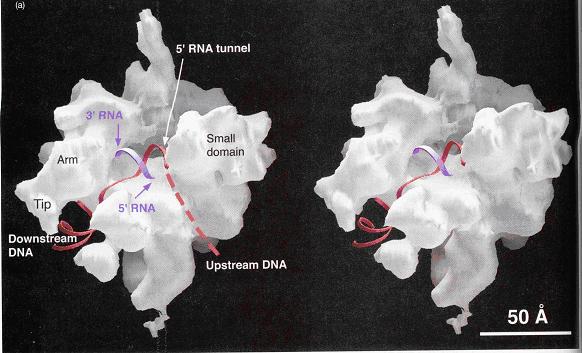
Image of yeast RNA polymerase II (Weaver 2002, p276)
A 25-Ǻ channel goes through RNAP with mainly Rpb1
on one side and Rpb2 on the other (Weaver 2002). The opening of the
channel is composed of Rpb1, Rpb9, and the Rpb5 subunit, which acts to
grasp DNA (Weaver 2002). Studies by Roger Kornberg and colleagues
show that DNA and RNA bind in the central cleft between Rpb1 and Rpb2 subunits.
The surface charge distribution of yeast at 2.8 Ǻ resolution shows that
hundreds of positively charged amino acid side chains line the cleft appropriately
for interaction with negatively charged DNA and RNA (Kornberg 2001).
Duplex DNA enters RNAP II and unwinds 3 bases before the active site.
The template strand makes a 90˚ bend and points downward toward the floor
of the cleft for readout at the active site (Kornberg 2001). The
ribonucleotide on the RNA chain is then base paired to the DNA strand forming
a RNA-DNA hybrid (Kornberg 2001). The structure determination of a
crystallized transcribing RNAP proved that the RNA and DNA interact in the
RNAP cleft (Kornberg 2001).
The Rpb2 subunit of RNAP forms a wall on the upstream
side of the active site that deters DNA from extending upstream with an
orientation like downstream DNA (Weaver 2002). The RNA-DNA hybrid
upstream from the active site is at an angle (dashed line) to the downstream
DNA (Weaver 2002). The pores depicted allow exit of the growing RNA oligomer
and hold a sliding clamp (composed of Rpb1, Rpb2, and Rpb6 subunits) on
the DNA improving processivity (Weaver 2002).
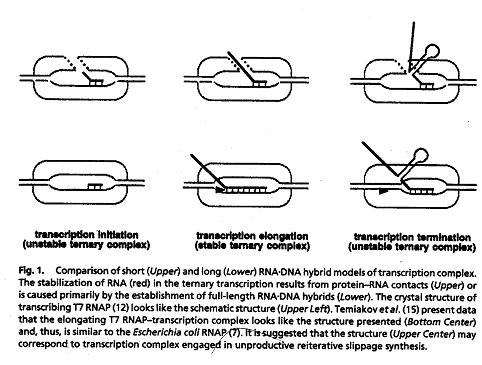
DNA-RNA hybrid model of transcription complex (Severinov 2001,p6)
HOME to RNA Pol II in Eukaryotes and Prokaryotes
BACK to GTF
NEXT to Comparison bacterial vs. eukaryotic RNAP