Dna B Helicase
|
Structure |
|
|
Character |
|
|
Function |
|

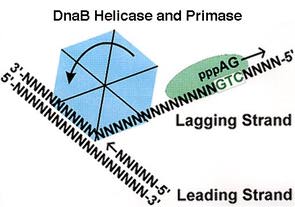
The unwinding of double stranded DNA using Dna B helicase. This picture was gathered from the http://chem-mgriep2.unl.edu/replic/Helicase.html .
Dna B is a protein from the Dna B gene. This protein is a main constituent for the prokaryotic primosome complex. This complex leads to the replication of DNA.
A primosome is a complex main of two proteins. Dna B and Dna G are the two proteins that cause replication to begin.
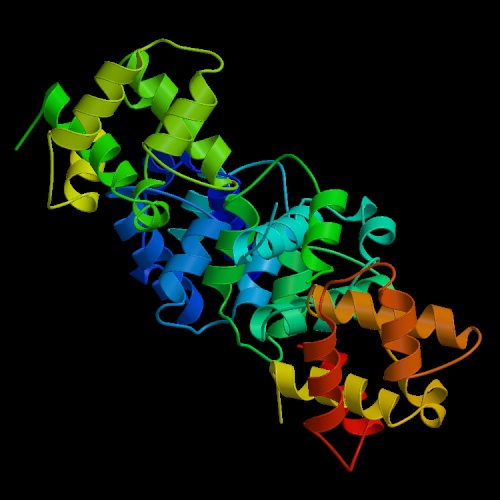
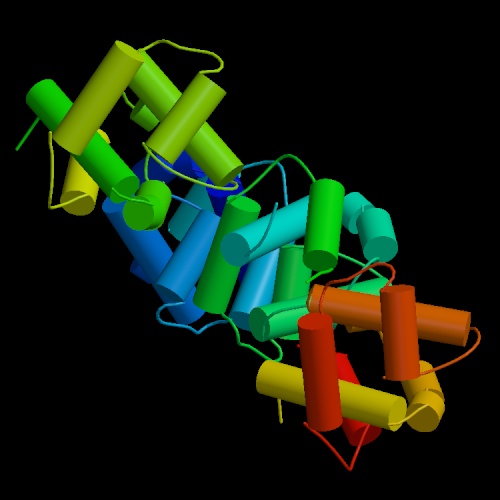
These are both pictures of the N-Terminal Domain Of DNA Replication Protein Dna B, the one on the right shows it in ribbon form and the one on the left shows it in cylinder form. These pictures were obtained from http://www.helicase.net/struct.htm#DNAB.
Discovery
In 1986, Jonathan LeBowitz and Roger McMacken discovered that Dna B protein was a helicase. Single stranded DNA of about 1000 base pairs were radio labeled with phosphate. The phosphate tracked the amount of rNTP’s used by the Dna B protein. This seemed logical since Dna B was already proven to be an ATPase. This function of Dna B helps push replication forward with the help of single stranded binding proteins(SSB).
Kornberg and colleagues discovered the importance of Dna B when they assayed a single stranded Φx174 phage DNA. When it produced ds DNA, Kornberg realized it needed primase synthesis before replication. So the pol III enzyme was used by DNA replication but other proteins are needed for primer synthesis. That is how they discovered the importance of Dna B for DNA replication.
Functions of Dna B
DNA replication begins at the oriC. This is the site where proper replication occurs. The site is 245 base pairs long. This origin contains 4 nine-mers which are the binding sites for a protein called Dna A. The Dna A protein controls the binding of Dna B to the origin. This binding occurs when Dna A initiates the melting of 3 13-mers on the other end of the origin site. This melting of the base pairs causes the Dna B to bind to the origin. Although there is still a problem with Dna B finding the site. So Dna C binds to Dna B to help deliver the complex to the open complex or DNA melting region of origin. After Dna B binds to the 13-mer melted region, this stimulates the binding of Dna G (primase). Thus completing the primosome. The Tao subunit of the DNA polymerase stabilizes and stimulates the Dna B helicase. As the Dna B is stabilized, the protein begins to unwind the dsDNA as it hydrolyzes ATP. The hydrolyzed ATP causes a slight structure change which helps in the ability to connect with the correct sites and move the ssDNA in one direction. Once replication has started there are two main functions the Dna B helicase serves
1. Consistently needs to bind for priming on the lagging strand because the lagging strand produces Okazaki fragments
2. Dna B unwinds the parental DNA to give the pol III templates for the leading and lagging strand, this occurs in the direction of 5’ to 3’. This is the same direction as the replication fork is moving
References:
Cox, M. and Nelson, D. (2000) Lehninger Principles of Biochemistry. Pg 941-946.
"DNAB Helicase." http://chem-mgriep2.unl.edu/replic/Helicase.html. (June 26, 2000).
Fass, D., Bogden, C. E., Berger, J. M.: Crystal Structure of the N-Terminal Domain of the Dna B Hexameric Helicase Structure (London) 7 pp. 691 (1999).
Kornberg, A and Baker, T. (1992) DNA Replication. (Ed. 2). Pp. 283.
LeBowitz, J and McMacken, R. (1986) The Journal of Biological Chemistry. 261, pp.4738-4748.
Snustad and Simmons. (2000) Principles of Genetics. Pg 255-280
Weaver, R. F. (1999) Molecular Biology. Pp. 668- 669.