The transcription factor NF-kB was discovered over 15 years ago, yet, owing to its wide range of important cellular roles, it remains an area of interest and current research. This versatile molecule was originally encountered as a factor necessary for the transcription, not of interferon, but a different immune response macromolecule--immunoglobin. Specifically, a 1986 paper first described NF-kB as a factor regulating the formation of the kappa variety of the light chain (one of two subunits from which immunoglobin is built). Discovered in the "B cell" white blood cell type, NF-kB is the Nuclear Factor for the k immunoglobin light chain in B cells. Since it was first observed, NF-kB has been found to play an active roll in inflammatory responses, cellular growth, and apoptosis as well as being present in diseases such as cancer, arthritis, asthma, and others.
NF-kB is the most widely encountered member of a family of transcription factors, the dimers of Rel proteins. The Rel family members contain a structurally similar motif, the Rel homology region (RHR, also written simply RH). The RHR is responsible for locating the binding sequence on DNA, dimerization with other Rel proteins around the DNA, and interactions with inhibitor proteins in the cytoplasm. Rel proteins are utilized by many eukaryotic organisms and the RH domain is conserved throughout them. While all Rel proteins share structural similarity, they can be further divided into two classes based upon the sequence on the C-terminal side of the RHR. Initially dormant, the first class of proteins contain a long chain of repeats that inhibits their function. p105, a member of this class is the precursor to one of NF-kB's monomers. Upon activation, the repeat chain is cleaved and P105 becomes p50. The second class of Rel proteins contain a transcription activator region on the C-terminus side of the RHR. NF-kB's second monomer, p65 (also known as RelA), is a member of this group. The p65 subunit of NF-kB is thought to have an additional synergy domain (in addition to the activation domain), which is necessary for the enhanceosome's success.
In addition to the heterodimer, p50/p65, homodimers of the Rel proteins exist also. (The structures of the homodimers were actually determined several years before a structure for the NF-kB heterodimer was elucidated.) p50 homodimers do not induce transcription. They are thought to be used as post-induction repressors, competing with NF-kB following transcription activation subsequent to a viral invasion.
NF-kB functions as a fast messenger. Under normal circumstances, it is sequestered in the cytoplasm by an inhibitor protein. Additionally, the p50 subunit is in the inactive elongated p105 state. Once a viral infection is recognized, the inhibitor is phosphorylated, releasing the complex, and the repeated chain is cleaved. Because the proteins originate in the cytoplasm already assembled and proceed to the nucleus where they act as the transcription factor, the cellular message is very fast.
Together, p50 and p65 dimerize around a 10 base pair region known as a kB site, forming the NF-kB transcription factor. The binding sequence is 5'-GGGRNYYYCC-3' (where R is a purine, Y is a pyrimidine, and N is an unspecified base).
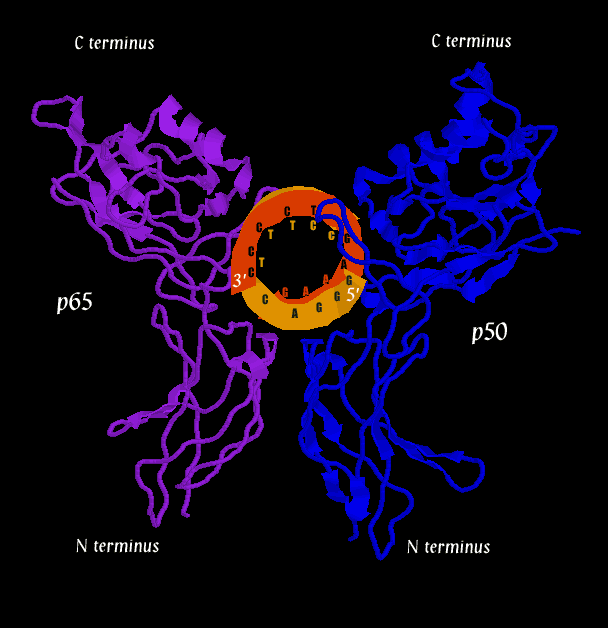
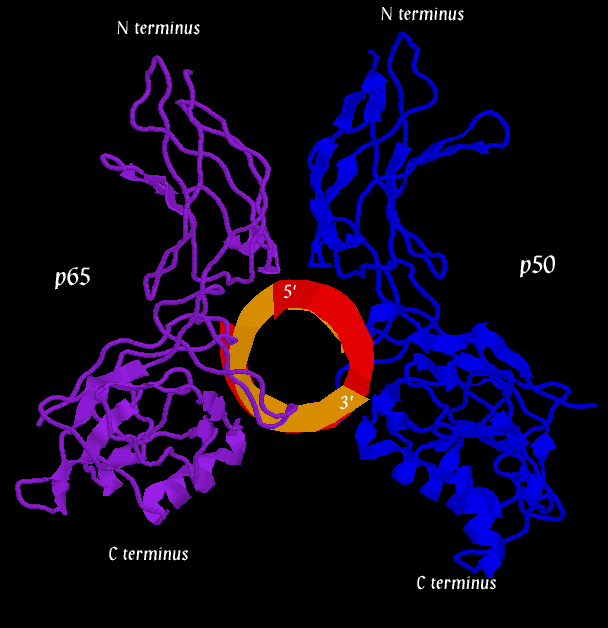
Below are three pictures illustrating the general structure of the assemblage. They show the NF-kB dimer binding to a kB sequence upstream of the immunoglobin light chain gene in mouse cells (Mus Musculus).
The second figure (the side view) helps to illustrate how, together, p50 and p65 can "feel out" the major groove over nearly one whole helical repeat by interacting in the major groove on opposite sides of the strand. In the first figure (top view), a loop of the p50 monomer (blue) can be seen winding into the DNA (at the 1 o'clock to 3 o'clock region). This loop is again visible in the second figure (side view) near the 5' end of the orange DNA strand. It is this loop that interacts with a 5-base pair sequence, 5'-GGGRN-3'. A similar loop can be seen clearly in the third figure (bottom view), in which a region of the p65 monomer makes contact with the DNA. The loop is also visible in the second figure (side view), although it is tucked in the back. The p65 loop provides recognition for a 4-base pair sequence, 5'-YYCC-3'.

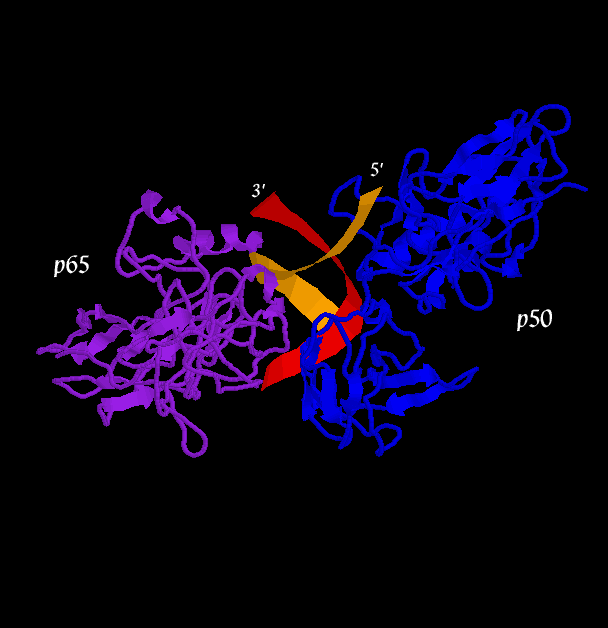
One final view highlight the spatial positions of the interacting protein residues.
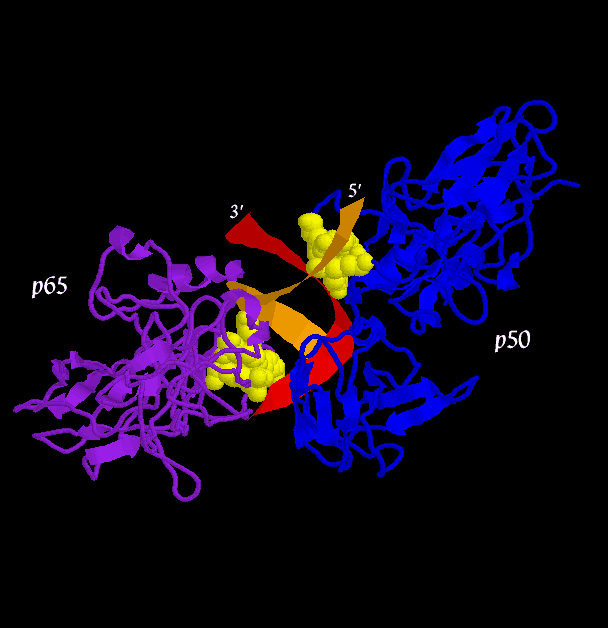
The mechanism of base pair recognition is shown for the p50 loop to give a sense of the interactions involved. What is not shown in this mechanism are the many interactions with the protein backbone and the p65 monomer.
While p50 and p65 homodimers exist, the scientists that elucidated the NF-kB crystal structure pointed out one special interaction between an Asp residue on p50 and an Asn on p65. In the homodimers, these groups react unfavorably, but in the heterodimer, they promote bonding.
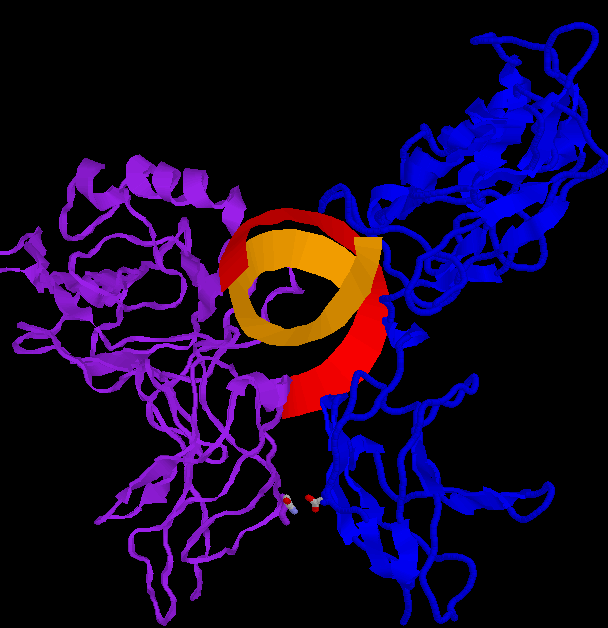
The structures used in creating all of the images come from Chen, F. E., et al. The PDB file (ID= IVKX) can be found here. The NF-kB comes from Mus Musculus.
An additional structural study involving inhibitor action on NF-kB done
by Huxford, T. et al. provides a second useful PDB file (ID= 1IKN).
The proteins were isolated from Mus Musculus and Homo Sapiens.
A suggested review article is
Ghosh, S. et al., which links to extensive online resources.