The jun gene codes for sequence-specific trans-acting factors which act as
functional sites on the AP-1 transcriptional complex. This gene codes for several
transcriptional factors via heterogeneous initiation and polyanylation sites.
The jun gene is about 2.7 kilobases and it is an intronless gene that is
located to the chromosomal 1 p31-32 region. The jun gene is part of a
highly conserved gene family found in different organisms such as v-jun from
avian sarcoma virus 17 and GCN4 from the yeast transcription activator.
C-jun was first believed to encode AP-1; however, several experiments
have determined that jun encodes the activation site of AP-1 rather than the
whole AP-1 complex. That is, AP-1 is composed of many other proteins,
which may enhance or repress the transcriptional machinery. Angel
et al. determined that there is a TPA-responsive element (TRE) about 9 base
pair motif, which is recognized by a trans-acting factor and enhances
the transcription machinery. They termed this sequence specific DNA
binding protein as the TRE-binding factor.
The jun gene contains a polyadenylation signal sequence downstream of
its 3' end and is very A-T rich at its 3' untranslated region. On its 5' end,
the jun gene has a short sequence such as 5' GATAAG and 5' TATTTTTA, which
is located 30 bp upstream of the initiation site and is proposed to be
homologous to the conventional TATA box. Also, the 5' untranslated end has
very high G-C content. Angel et al. suggest that a consensus CAAT box
and GC box that are further upstream of these sequences are potential
promoter elements. These findings and relation to A-T rich sequences on
the 3' untranslated region on c-fos suggested that the DNA binding domain
of the jun product is located on it C-terminus end. This C-terminus end
contains a leucine repeat region, which can interact with a similar region on
another protein to form a leucine zipper dimer. The dimer
binds DNA, thereby inducng the transcription machinery. In the case of the
INF- enhanceosome, c-Jun forms a heterodimer with
ATF-2, as shown below.
enhanceosome, c-Jun forms a heterodimer with
ATF-2, as shown below.
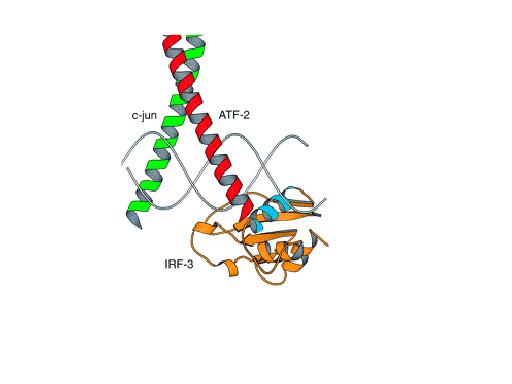
(adopted from:www.fas.harvard.edu/~mcb155)
The jun gene codes for cJun, JunB, and JunD. The cJun protein is the polypeptide that is found in the active site of the AP-1 complex in cellular organisms. The finding followed the isolation of vJun oncogene from Avian sarcoma virus 17 by Vogt and coworkers. Since AP-1 recognizes the TRE consensus sequence and vjun and cjun encode the proteins in AP-1 that also bind TRE, it was determined that the jun product is part of the functional domain of the AP-1 complex. Jun is found in both homodimer and heterodimer associations, each form having different binding activities and functions. Dimerization occurs at the leucine repeat region found on the protein C-terminus end for both the heterodimer (formed, for example, with the Fos protein, which is another major component of the AP-1 active site) and for the homodimer (formed by two Jun proteins). The leucine repeats at every seventh amino acid on each monomer and interacts via hydrophobic interactions and by formation of salt bridges, which help stabilize the dimer. The interactions form the extensive a-helical leucine zipper, as in the case of the Jun:Fos heterodimer. Upstream of the leucine repeat region, there is the basic region that contains abundant positively charged residues. This region functions as the DNA-binding site domain. The basic region on all Jun proteins is highly conserved. Also, there are clusters of negatively charged residues on the N-terminus end of the Jun protein that are important in transcriptional activation. The major role of heterodimeric Jun is to bind DNA and initiate transcriptional activation by the AP-1 complex in response to extracellular stimulation. The heterodimer initiates transcriptional activation by dephosphorylation of specific serine and threonine residues. The serine and threonine residues are located upstream of the basic region and are also conserved in all Jun products. Dephosphorylation of these residues greatly increases the DNA-binding affinity of Jun and phosphorylation by the glycogen synthase kinase 3 (GSK-3) decreases the DNA binding activity on the TRE site of AP-1. Thus, phosphorylation and dephosphorylation of Jun on the AP-1 complex regulates initiation of transcription, making Jun a positive and negative regulator of AP-1 activity. The other Jun products, JunB and JunD enhance repressive activity and control the amount of the Jun protein present via extracellular stimulation, that is, they regulate cJun. Furthermore, cJun and its counterparts become oncogenic when the phosphorylated residues undergo mutatons leading to constant activation of cJun.
extracted from 1A02 (chain J), retrieved from the Protein Data Bank:
(From Homo Sapiens)

- Angel, P., Karin, M. , The role of Jun, Fos and the AP-1 complex in cell, Biochim. et Biophys. Acta (1991)
- Kallunki, T., Su, B., Tsigelny, I., Sluss, H.K., Derijard, B., Moore, G., Davis, R. and Karin, M. 1994. JNK2 contains a specificity-deermining region responsible for efficient c-Jun binding and phosphorylation. Genes and Development 8, 2996-3007
- Angel P, Allegretto A E., Okino S T., Hattori K, Boyle W J., Hunter T, Ka rin M. Oncogene jun encode a sequence-specific trans-activator similar t o AP-1. Nature 1988: Vol. 332: 166-170
- Angel P, Imagawa M, Chiu R, Stein B, Imbra R J., Rahmsdorf H J., Jonat C, Herrlich P, Karin M. Phorbol Ester-inducible genes contain a common cis element recognized by a TPA-modulated Trans-acting factor. Cell 1987: V ol 49: 729-739
- Bohmann D, Admon A, Turner D.R,, Tyian R. Trancriptional Regulation by t he AP-1 family of enhancer-binding proteins: A nuclear target for signal transduction. Cold Spring Harb. Symp. Quant. Biol. Vol. LIII. 1998: 695 -700.
- Curran T, Franza B R. Jr. Fos and Jun: The AP-1 connection. Cell 1988: Vol. 55, 395-397
- Engel James Douglas. Meticulous AP-1 factors. Nature 1994: Vol. 367: 51 6-5177
- Junius FK, O'Donoghue SI, Nilges M, Weiss AS, King GF. High resolution N MR solution structure of the leucine zipper domain of the c-Jun homodimer : J Biol Chem 1996 Jun 7;271(23):13663-7
- Kazue Hattori, Peter Angel, Michelle M. Le Beau, and Michael Karin. Stru ctural and chromosomal localization fo the functional intronless human JU N protooncogene: Proc. Natl. Aced. Sci. USA 1988 Dec Vol. 8: 9148-9152
- Chen, L. et al., Structure of the DNA-binding Domains from NFAT, Fos and Jun bound specifically to DNA, Nature, 392, p.42 (1998)