High Mobility Group Proteins
High Mobility Group (HMG) proteins are a class of non-histone chromosomal species, which share the common property of rapid migration in PAGE gels. Represented in all four eukaryotic kingdoms, and ubiquitous in mamalian cells, HMG proteins were first isolated from calf thymus chromatin and characterized in 1973 (1). The first isolated species, named HMG-1 and HMG-2 attracted interest due to their high content of both basic and acidic amino acids. HMG-1 and HMG-2 are now known to recognize a DNA binding motif known as the "HMG-box", and are classified in the HMGB protein superfamily.Additional species were subsequently isolated and numbered according to their locations on an electrophoretic gel. All but three, namely HMG-14, HMG-17 and HMG-20 were later determined to be either non-chromosomal, or resulting from proteolysis of the other species (2). HMG-20 was later identified as Ubiquitin, and is no longer classified with the HMG proteins (3). The HMG-14 and HMG-17 proteins were initially distinguished from the HMG-1 and HMG-2 proteins by their lower molecular weights of 10-12 kD, compared to the 28 and 27 kD weights of the earlier two species (4). HMG-14 and HMG-17 are now recognized as nuclosome binding domain (NBD) proteins, and belong to the HMGN protein superfamily.
HMG-I(Y) Proteins
Two new low molecular weight (10-12 kD) "HMG-like" proteins were isolated from human cells in 1983 (5), and designated HMGI and HMGY. The HMGI protein was also detected in mouse cells (6,7). The I and Y are now known to be isoforms of the same protein (8). The HMG-I(Y) sequence, structure and expression patterns have now been chracterized for mouse (9) and for human cells (10). These proteins are of particular interest because they exhibit specific binding to the minor groove of AT-rich sequences of B-form DNA, via highly conserved peptide motifs termed "AT hooks", thereby stabilizing the B-form and facilitating the binding of transcription factors in the opposing major groove. These species demonstrate the ability to recognize distorted DNA, and to bend, unwind, supercoil and generally reshape bound DNA substrate, in vivo and in vitro. AT-hook motif binding proteins include the HMGI and HMGY isoforms as well as the HMGI-C homologue. The AT-hook motif proteins are now classified in the HMGA protein superfamily.
The HMG-I(Y) function as architectural proteins in the
INF-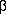 enhanceosome. Upon binding to
the DNA, they remove the intrinsic bend of the double helix, allowing
the binding of transcriptional activators.
HMG-I(Y) binds the INF-
enhanceosome. Upon binding to
the DNA, they remove the intrinsic bend of the double helix, allowing
the binding of transcriptional activators.
HMG-I(Y) binds the INF-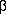 enhancer
sequence at four locations, on two different Positive Regulatory Domains:
PRD-II and PRD-IV. The second binding domain of one HMGI protein binds
cooperatively with the NF-
enhancer
sequence at four locations, on two different Positive Regulatory Domains:
PRD-II and PRD-IV. The second binding domain of one HMGI protein binds
cooperatively with the NF- B element
at PRD-II, while the associated third binding domain binds precisely
one helix turn downstream in the NRDI region. The PRD-II is intrinsically
bent by 20 degrees. The binding of
HMGI and NF-
B element
at PRD-II, while the associated third binding domain binds precisely
one helix turn downstream in the NRDI region. The PRD-II is intrinsically
bent by 20 degrees. The binding of
HMGI and NF- B
induces a bend of 26 degrees in the opposite direction, resulting in an
overall change in bending of 46 degrees, and facilitating further interaction
amongst the enhanceosome components. A second HMGI protein is known to
cooperatively bind either side of the PRD-IV region, along with the
ATF-2/c-Jun heterodimer. Here again, binding removes the intrinsic bend of
the DNA by introducing non-native bending.
B
induces a bend of 26 degrees in the opposite direction, resulting in an
overall change in bending of 46 degrees, and facilitating further interaction
amongst the enhanceosome components. A second HMGI protein is known to
cooperatively bind either side of the PRD-IV region, along with the
ATF-2/c-Jun heterodimer. Here again, binding removes the intrinsic bend of
the DNA by introducing non-native bending.
The synergetic activation of the INF-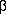 is
highly dependent on the three-dimensional conformation of the enhanceosome
complex. HMGI plays a vital role in this context. It has been determined
that the introduction of helical phasing mutations between PRD-II and PRD-IV
significantly reduces the enhancement of interferon expression. Thus, the
binding of HMGI proteins to the minor groove on the same face of the
enhancer region is essential in bringing the transcription factors on the
opposite face into close contact. The importance of the cooperative binding
of HMGI and the ATF-2 component is clearly demonstrated by experiments
replacing ATF-2 with an isoform ATF2192. While the
latter form can bind to PRD-IV, and is capable of participating in the
enhanceosome complex, overall transcription is severly diminished as it
cannot interact with the HMGI.
is
highly dependent on the three-dimensional conformation of the enhanceosome
complex. HMGI plays a vital role in this context. It has been determined
that the introduction of helical phasing mutations between PRD-II and PRD-IV
significantly reduces the enhancement of interferon expression. Thus, the
binding of HMGI proteins to the minor groove on the same face of the
enhancer region is essential in bringing the transcription factors on the
opposite face into close contact. The importance of the cooperative binding
of HMGI and the ATF-2 component is clearly demonstrated by experiments
replacing ATF-2 with an isoform ATF2192. While the
latter form can bind to PRD-IV, and is capable of participating in the
enhanceosome complex, overall transcription is severly diminished as it
cannot interact with the HMGI.
Recruitment of the ATF-2/c-Jun and NF- B dimers
is an essential function of HMGI in activation of the INF-
B dimers
is an essential function of HMGI in activation of the INF- gene. It has been shown that addition of ATF-2/c-Jun and
NF-
gene. It has been shown that addition of ATF-2/c-Jun and
NF- B in vitro, in the absence of HMGI results
in only weak transcription, whereas the inclusion of HMGI increases
transcription levels almost 30-fold. It has been further demonstrated that
HMGI plays a central role in activating the INF-
B in vitro, in the absence of HMGI results
in only weak transcription, whereas the inclusion of HMGI increases
transcription levels almost 30-fold. It has been further demonstrated that
HMGI plays a central role in activating the INF-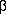 transcriptional switch in the following manner. Acetylation of specific
HMGI residues (Lys 71) stabilizes the enhanceosome complex,
potentiating transcription,
whereas the acetylation of other specific HMGI residues (Lys 65)
destabilizes the
complex and terminates transcription.
transcriptional switch in the following manner. Acetylation of specific
HMGI residues (Lys 71) stabilizes the enhanceosome complex,
potentiating transcription,
whereas the acetylation of other specific HMGI residues (Lys 65)
destabilizes the
complex and terminates transcription.
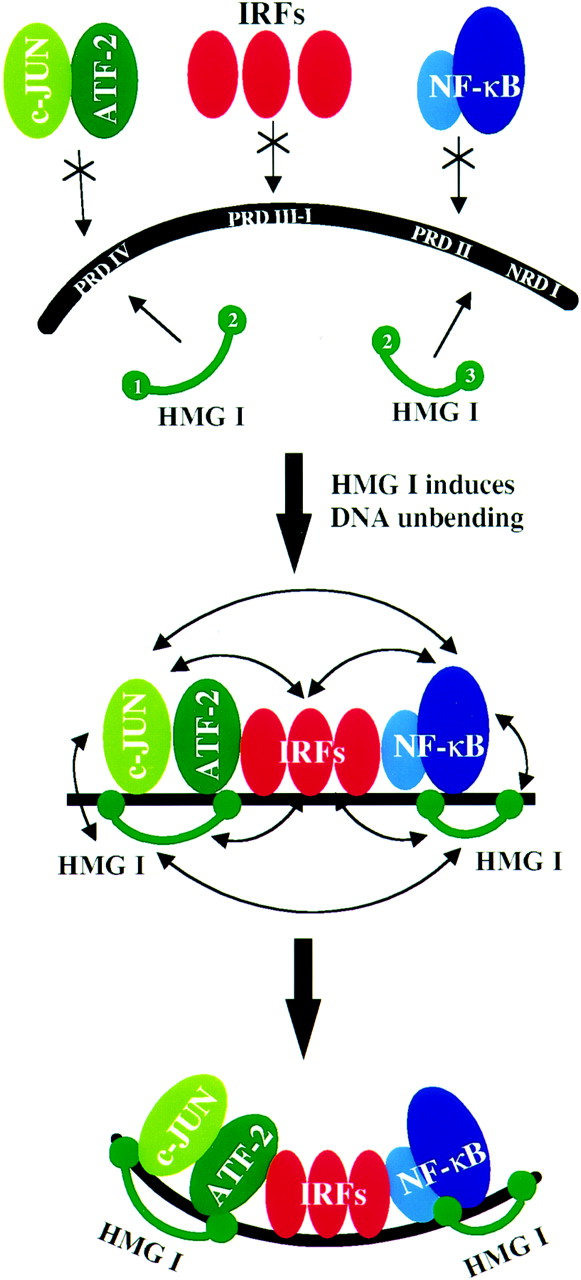
(Source: Yie, J. et al. (1999) The role of HMG I(Y) in the
assembly and function of the IFN- enhanceosome, EMBO J., 18, p.3086)
enhanceosome, EMBO J., 18, p.3086)
The HMG-I(Y) proteins include three binding domains, imaginatively dubbed
"first", "second" and "third".
The stucture of this protein has not yet been fully resolved.
However, the solution structure of segments of the HMG-I(Y)-DNA complex
have been determined via multidimenstional NMR,
as shown below. These resolved stuctures
consist of the second and third DNA binding domains (residues 51-90),
along with the associated DNA dodecamer containing the PRDII site of the
interferon- promoter.
promoter.
PDB files from the
Protein Data Bank:
(All structures from Homo Sapiens)
first binding domain - no PDB file available
second binding domain -
NMR, minimized average structure, PDB id:
2EZD
NMR, 35 structures, PDB id:
2EZE
third binding domain -
NMR, minimized average structure, PDB id:
2EZF
NMR, 35 structures, PDB id:
2EZG
Images generated from PDB files:
Second binding domain, view facing minor groove:
Second binding domain, view along DNA helix axis:
Third binding domain, view facing minor groove:
Third binding domain, view along DNA helix axis:
- Goodwin, G. H., Johns, E. W., Isolation and Characterisation of Two Calf-Thymus Chromatin Non-Histone Proteins with High Contents of Acidic and Basic Amino Acids, Eur. J. Biochem, 40, 215-219 (1973)
- Johns, E. W. (ed.), The HMG Chromosomal Proteins, Academic Press, New York (1982)
- Goodwin, G. H., et al., Biochim biophys acta, 519, 233 (1978)
- Einck, L. and Bustin, M., The Intracellular Disstribution of the High Mobility Group Chromosomal Proteins, Experimental Cell Research, 156, 295-310 (1985)
- Lund, T. et al., On the Presence of Two New High Mobility Group-like Proteins in HeLa S3 cells, FEBS lett.,152, 163-167 (1983)
- Lund, T. et al., FEBS lett.,180, 275-279 (1985)
- Elton, T. S. and Reeves, R., Purification and Postsynthetic Modifications of Friend Erythroleukemic Cell High Mobility Group Protein HMG-I, Anal. Biochem., 157, 33-62 (1986)
- Lehn, D. A. et al., Biochem. Int., 16, 963-971 (1988)
- Johnson, K. R. et al., Complete Murine cDNA Sequence, Genomic Structure, and Tissue Expression of the High Mobility Group Protein HMG-I(Y), Journ. Biol. Chem., 263, 18338-18342 (1988)
- Friedmann, M. et al., Organizaion, Inducible-expression and Chromosome Localization of the Human HMG-I(Y) Nonhistone Protein Gene, Nucl. Acid Res., 21, 4259-4267 (1993)
- Reeves, R., Structure and Function of the HMGI(Y) Family of Architectural Transcription Factors, Environ. Health Persp., 108, Suppl. 5 (2000)
- Kim, T. K. & Maniatis, T., The Mechanism of Transcriptional Synergy of
an In Vitro Assembled Interferon-
 Enhanceosome,
Mol. Cell, 1, 119-129 (1997)
Enhanceosome,
Mol. Cell, 1, 119-129 (1997) - Yie, J. et al., The role of HMG I(Y) in the Assembly and
funcion of the INF-
 Enhanceosome,
EMBO J., 18,
3074-3089 (1999)
Enhanceosome,
EMBO J., 18,
3074-3089 (1999) - Munshi, N. et al., Coordination of a Transcriptional Switch by HMGI(Y) Acetylation, Science, 293, 1133-1136 (2001)
- Huth, J. R., Bewley, C. A., Nissen, M. S., Evans, J. N., Reeves, R., Gronenborn, A. M., Clore, G. M., The Solution of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Biol pp., 657 (1997)