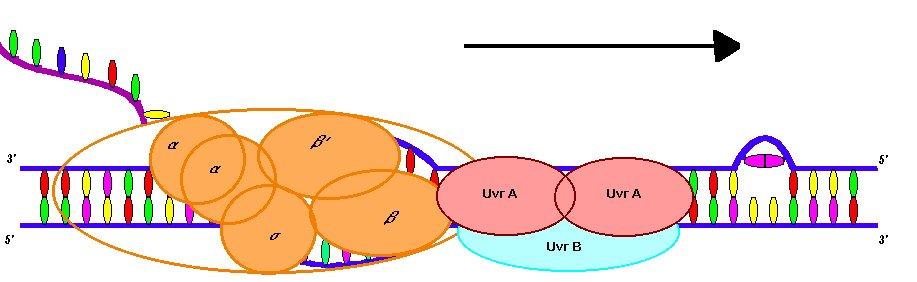
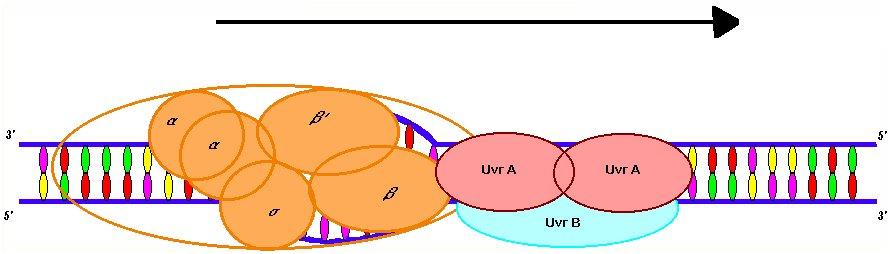
The UvrA2B complex is part of the prokaryotic nucleotide-excision repair (NER) cascade. NER is the mechanism responsible for removing non-specific DNA damage. UvrA is responsible for damage recognition via helix-turn-helix and polyhinge domains (Ahn and Grossman, 1996). It has been proposed that damage recognition occurs through helix distortion rather then direct read-out (Voet, Voet, and Pratt, 1999).
The binding of UvrA to damaged DNA is about 103 and 104 times greater then undamaged DNA (Ahn and Grossman, 1996). UvrB is responsible for DNA binding and has endonuclease activity when paired with UvrC (Snustad and Simmons, 2000). UvrB is associated with a sharp bend created in the DNA, which facilitates the release of UvrA and the binding of UvrC (Selby and Sancar, 1993).
The RNA polymerase open complex facilitates the loading of the UvrA 2B complex onto a single stranded section of the DNA. In fact, the complex binds preferentially to promoter sequences in the open complex (Ahn and Grossman, 1996). This phenomenon has been hypothesized to also occur in eukaryotes where transcription-coupled repair is observed to act on a gradient with the greatest degree of repair occurring near the promoter region of the gene. This has been explained by increased concentrations of repair enzymes near the initiation site due to associations with RNA polymerase and its transcription factors (Tu, Tornaletti and Pfeifer, 1996). In prokaryotes, this concentration is most likely brought about by an affiliation of the UvrA 2B complex for RNA polymerase. Cross-linking experiments with DSP (dithiobis-succinimidylpropionate) and immunoprecipitation with antibodies to UvrA indicate a physical association between the b subunit of RNA polymerase and UvrA (Lin, Kovalsky, and Grossman, 1998). Since the loading of UvrB onto DNA is the rate-limiting step in NER, the preferential loading of the UvrA2 B complex by the RNA polymerase complex is the first step of linking transcription to repair (Selby and Sancar, 1993).

After loading UvrA2B moves along the non-template DNA strand in search of kinkable errors in the template strand. UvrA2B has specific ATP-dependent 5’ to 3’ helicase activity. Therefore, the complex must bind to the non-template strand to prevent collisions with RNA polymerase (Ahn and Grossman, 1996). It is not proven whether UvrA and the b subunit of RNA polymerase are associated during the elongation or only during the initiation phase of transcription. Consequently, it is unknown whether the UvrA2B moves with or speeds ahead of the RNA polymerase complex during translation (Lin, Kovalsky, and Grossman, 1998).