ORC = Origin Recognition Complex
Mcm = Minichromosome maintenance proteins
Cdc6 = cell-division-cycle
Cdk = cyclin-dependent kinases
BACKGROUND
The heterohexameric origin recognition complex (ORC) is the eukaryotic analog to prokaryotic and viral initiator proteins (Bielinsky 2000). As a DNA replication initiator protein complex, ORC fulfills three basic functions:
1) Unwinding the origin by using helicases (in eukaryotes it is believed that
Mcm’s function as a helicase)
2) Guiding the replication machinery to the origin.
3) Linking cell cycle control to origin activation (Bielinsky 2000).
The ORC can be thought of as a molecular machine in that it recruits other proteins (such as cell cycle-regulated Cdc6p) to the origin at the crucial time and in the key sequence for replication to begin. DNA binding of the ORC requires ATP (Bell 2002).
ORC is composed of 6 subunits, ORC 1p-6p. Generally, the function and size of the ORC subunits are conserved across many eukaryotic genomes, however, which unit actually contacts the DNA is distinctly different. The most widely studied origin replication complex is that of Saccharomyces cerevisiae (yeast) which binds to the ARS (autonomously replicating sequence). The origin replication sites in higher eukaryotes are comprised of similar A-T rich regions and may have their evolutionary history in the ARS site of yeast (Bielinsky 2000).
In yeast, the pre-Replication Complex (pre-RC) is composed of the ORC, Cdc6p, and Mcm2p-7p. ORC binds to A-T rich regions of the DNA, and there is some evidence that additional interactions with a B1 domain are vital to ORC binding affinity (Bell 2002). In some organisms, an additional factor, Cdt1, is necessary for loading Mcm onto the chromatin. The ORC-Cdc6 complex may couple ATP hydrolysis to conformational changes in the Mcm complex. During late mitosis when Mcm binds, it has been suggested that ORC and Cdc6 may no longer be required and may be removed (Kelly 2000). In mammalian cells, the order of complex formation is still being debated. It has been shown that ORC 2p-5p make stable complexes, but ORC 1 is difficult to detect in vivo (Fujita 2002).
THE INITIATION COMPLEX
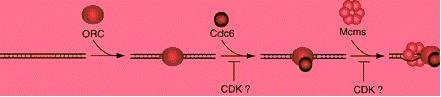
the MCM proteins. CDKs inhibit the formation of the initiation complex, in part by blocking the expression,
nuclear localization, or activity of Cdc6/Cdc18. Possible points of regulation are indicated in the figure
(Kelly 2000).
ACTIVATION OF THE INITIATION COMPLEX
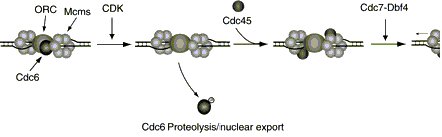
the initiation of DNA synthesis is triggered by the action of CDK and Cdc7 family protein kinases.
One consequence of CDK activity is the loading of Cdc45, DNA polymerase alpha (Pol alpha ) and
probably other proteins that function at the replication fork. One of the final steps in the initiation reaction
may be the activation of the helicase activity of the MCM complex, possibly mediated by the Cdc7
protein kinase (Kelly 2000).
THE SIX SUBUNITS
(Note: All Molecular Weight approximations are from Saccharomyces cerevisiae, however, most eukaryotic ORC subunits are similar in size to this yeast)
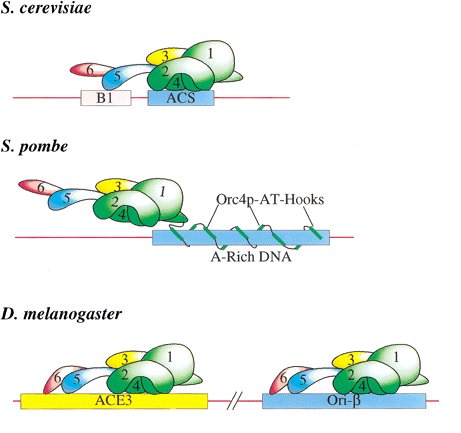
with Saccharomyces cerevisiae origins involves four of the six subunits (Orc1p, Orc2p, Orc4p, and
Orc5p) recognizing the 11-bp autonomous replicating sequence (ARS) consensus sequence (ACS)
as well as several less well-defined adjacent DNA sequences including a subset of B1 element
sequences (Bell 2002). Binding of Schizosaccharomyces pombe ORC (SpORC) to its cognate origins
involves a clearly distinct mechanism involving nine repeats of an AT-hook motif found uniquely at
the N-terminus of the S. pombe Orc4 subunit. A discrete binding site has not been identified,
however recent studies indicate that SpORC recognizes stretches of A-rich DNA. Less is known
about the sequences recognized by Drosophila ORC (DmORC), however, ATP- dependent binding
to both the ACE3 and ori-B elements of the third chromosome have been detected in vivo
and in vitro. Unlike ScORC, all six subunits of DmORC are required for DNA binding (Bell 2002).
ORC 1p
ORC 1p is a 120 kD subunit that forms close contact to the DNA and binds ATP (Bell 2002). When ORC binds to the DNA, the rate of hydolysis of ATP by ORC 1p changes. Thus, it is the central regulator of ORC function. Protein cross-linking experiments suggest that this subunit makes direct contact with the major groove of the DNA within 10 Å of the ACS site in yeast (Kelly 2000). Expression of ORC 1p is driven by elongation factor E2F (Fujita 10366).
ORC 2p
ORC 2p is a 72 kD subunit that also forms close contacts with DNA. It has been speculated that phosphorylation of ORC 2p inhibits re-initiation of replication through a feedback mechanism (Vas 2001). It, too, makes contact with the major groove of DNA near the ACS site. Expression of ORC 2p is cell growth independent (Fujita 2002).
ORC 3p
Little is known about this 62 kD subunit. It appears to lack any contact with the DNA, and may sit on top of the entire complex (Newlon 1993).
ORC 4p
ORC 4p is approximately 56 kD in size. This subunit is always in contact with origin DNA when the ORC complex is bound. It has been nicknamed the “anchor” because it tethers the origin recognition complex to the DNA (Kelly 2000). Protein linking experiments have revealed that ORC 4p contact the major groove of the DNA very close to the ARS site in yeast. In the S. pombe species, ORC 4p contains an N-terminal domain that binds to origin DNA in the absence of other subunits. In this domain, 9 copies of a hook bind to the minor groove of DNA in repeating 4-6 nucleotide A-T tracks (Kelly 2000). The binding of this subunit may be regulated by ATP.
ORC 5p
ORC 5p has a molecular weight of 53 kD. It is characterized by close interactions with the DNA replication origin sites and is thought to be regulated by ATP binding in much the same way as ORC 1p (Newlon 1993).
ORC 6p
ORC 6p is 50 kD and contains much of the same activities of ORC 2p except that it does not form close contacts with the DNA (Newlon 1993).