Primase (dna G)
E.coli DNA Primase (dna G) was discovered in 1972 by K. G. Lark. Kasper Zechel, Jean-Pierre Bouche and Arthur Kornberg pushed this discovery further when they uncovered the function of dna G through the use of two phage strains and the wild-type E.coli uninfected. The phage strains were G4, and G14. They assayed the modified E.coli for RNA polymerase activity. Zechel et al reported the procedures and results in a paper in 1975 published in The Journal of Biological Chemisty (pp.4684-4689).
The dna G protein works as part of a multi-enzyme complex with dna B in order to prime the strands of DNA. Dna G seeks out a 3'- GTC sequence and adds on the primer about 11 nucleotides3. This complex is known as a "primosome." The primosome stimulates primer synthesis by RNA polymerase. The primosome continually moves along the lagging strand in order to prime each Okazaki fragment, and only primes once on the leading strand. Okazaki is credited with the discovery of much of these facts because of his studies of DNA replication which lead him to his conclusion of a semidiscontinuous mechanism.
References:
- Lark, K. G. (1972) Nature New Biology. 240, pp.237-240
- Zechel, K et al. (1975) The Journal of Biological Chemistry. 250, pp. 4684-4689
- Kornberg, A and Baker, T. (1992) DNA Replication. (Ed. 2). Pp.282-283
- Weaver, R. F. (1999) Molecular Biology. Pp.666
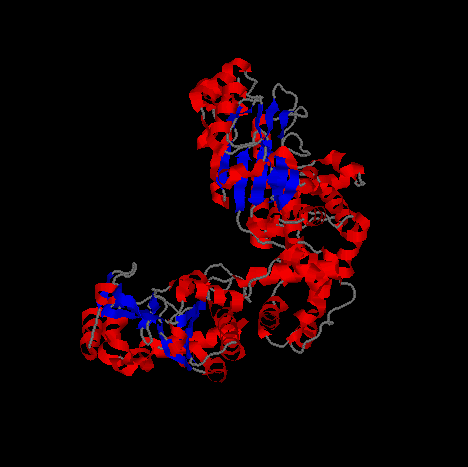
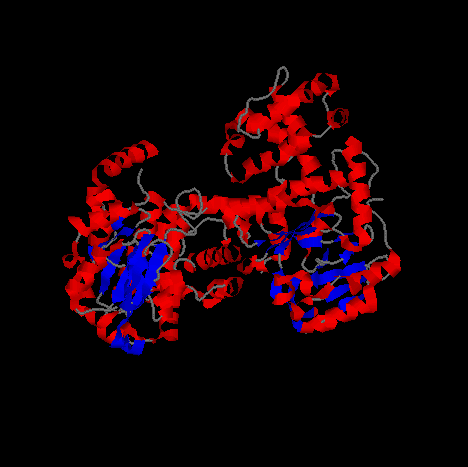
The images above and below are of Primase in a cartoon representation at different angles.
Helical segments of the protein are shown in blue and beta sheet regions are shown in red.
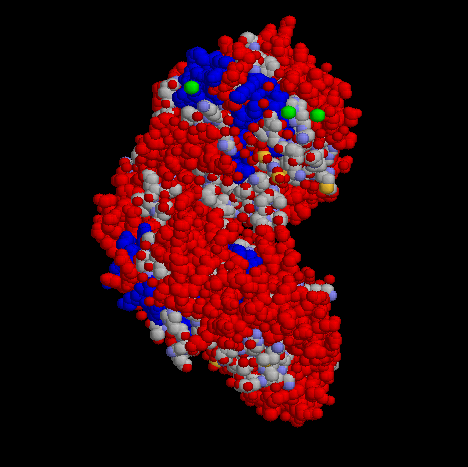
The above image is of Primase in a spacefilling representation in order to more easily see the three dimensional features of the compound.
Helical segments of the protein are shown in blue and beta sheet regions are shown in red.