Characterization of the a Subunit
In 1985 Hisaji Maki and Arthur Kornberg conducted experimentation to determine the characteristics of the a subunit. It was determined that the a subunit alone catalyzed the polymerization, but possesses neither 3'Æ5' nor 5'Æ3' exonuclease activities.
The asubunit protein was purified from an a subunit overproducing strain HB101. The specific activity of a subunit protein in the final preparation was found to be approximately the same as that of the polIII core. This would indicate that the subunit alone is responsible for the polymerization activity in the pol III core of the enzyme. The a subunit was found to have no exonuclease activity.
Maki, H., Kornberg, A. (1985) J. Biol Chem. 260. 12987-12992.
Characterization of e
In a study done by Richard H. Scheuermann and Harrison Echols in 1984 it was found that the e subunit is the subunit responsible for the editing 3'Æ5' exonuclease activity of DNA polymerase III. This was determined by overproduction and purification of the e subunit of the polIII enzyme. The dnaQ gene was overexpressed. It was determined that e was responsible for the exonuclease activity by noticing the lack of activity in defective dnaQ genes. It seems that e is capable of its exonuclease activity even in the absence of the a subunit suggesting that e functions independently of a. This functional separation may provide for regulation of exonucleolytic editing independently of polymerization, allowing for cellular control of replication fidelity.
Scheuermann, R., Echols, H. (1984) Proc. Natl. Acad. Sci. 81. 7747-7751.
Purification of DNA polymeraseIII holoenzyme into a, b, g, and d.
In 1977, Charles S. McHenry and Arthur Kornberg purified the DNA polymerase III holoenzyme from E. coli HMS-83, using as an assay, the conversion of phage G4 single-stranded DNA to the duplex replication form. It was found that the holoenzyme consists of at least four different subunits, a (140,000Da), b(40,000Da), g(52,000Da), and d(32,000Da).
The holoenzyme was obtained from Ammonium Sulfate Fractionation followed by Bio-Rex-70 Chromatography followed by Valyl Sepharose Chromatography and finally DEAE-Sephadex Chomatography. Th holoenzyme was then labeled with [35S] then denatured and electrophoresed on an SDS-polyacrylamide gel. Four protein bands were present. These bands were labeled a, b , g, and d.
The holoenzyme was then resolved by phosphocellulose chromatography into the b subunit and an agd complex. From this the a of the agd complex was selectively inactivated and dissociated by treatment with a zinc chelator, so that the gd complex could be purified as a distinct molecular species. This complex was then denatured and electrophoresed on an SDS-polyacrylamide gel. Two distinct proteins were observed.
From this experimentation, it was determined that there are at least four subunits that are required for conversion of a primed G4 single stranded circle to the duplex form. The a subunit was determined to be the polIII (it was later found that e and q are also part of the polIII) and the dnaE gene product. The b subunit is the copol III based on upon association with pol III stimulation of its activity on a long, single-stranded DNA template. The gd complex appeared to show no independent enzymatic activity but when combined with the a and b subunits reconstitute a holenzyme like activity.
McHenry, C., Kornberg, A. (1977). J. Biol Chem. 252. 6478-6484.
Identification of e and q as subunits of the DNA polymerase III holoenzyme.
In 1978 Charles S. McHenry and Weldon Crow determined that e and q were also subunits of the DNA polymerase III holoenzyme. In their experimentation, DNA polymerase was purified to homogeneitiy and the identification of these two additional subunits was possible.
DNA polymeraseIII was denatured and electrophoresed on an SDS-polyacrylamide gel. Three protein bands were identified and termed a (140,000Da), e (25,000Da), and q (10,000Da). The 140,000Da protein has been previously identified as a. All three proteins co-chromatographed in all purification procedures. All three proteins increased and decreased with polymerase activity upon gel filtration and phosphocellulose chromatography. From this it was concluded that a, e, and q are alls subunits of DNA polymerase.
When the polymer [3H]d(T)75[32P]d(C)8 was treated with polymeraseIII, the entire substate was digested meaning that the polymeraseIII contains a 3'Æ5' exonuclease which is specific for single stranded DNA. The presence of a 5'Æ3' was established by using the template [3H]d(T)75-[32P]d(C)8 to which excess oligo(dG)12-18 was annealed blocking the 3'Æ5' exonuclease. Polymerase III removed approximately equal amounts of thymidine and cytidine from the 3'-blocked template. When the 5' portion of the substrate is double stranded, it becomes inert to nuclease attack. This is demonstrated by annealing both poly(dA) and the oligo to the template. This demonstrates that DNA polymerase III also contains a 5'Æ3' exonuclease which must commence hydrolysis on a single strand of DNA but which can proceed into a double stranded region.
Identification of t as a Subunit of the DNA Polymerase III Holoenzyme
In 1981, Charles S. McHenry identified a new peptide subunit in DNA polymerase III holoenzyme through polyacrylamide gel filtration experimentation. He found DNA polymerase III' to be a complex of the a, e, q, and the newly assigned t subunit.
Solution containing the polymerase III' enzyme was applied to a 85mL Sephacryl s-300 column equilibrated in buffer. 1mL fractions were collected at a rate of 3.5ml/hr. Active DNA polymerase III' fractions were pooled. The pooled fractions were then denatured and electrophoresed on an SDS-polyacrylamide gel. Four major protein bands of 140,000, 83,000, 25,000, and 10,000 daltons were present. The largest and two smallest components were the previously known subunits of the polymerase III core, a,e, and q. The fourth, 83,000 dalton protein is not part of the core and is termed t.
The molecular weight of DNA polymerase III' was calculated from the Stoke radius and dedimentation coefficient by the method of Siegel and Monty. Gel filtration indicated that DNA polymerase III' is much larger than DNA polymerase III. The molecular weight of DNA polymerase III was found to be 160,000 and the molecular weight of DNA polymerase III' was found to be 410,000. From this it can be inferred that DNA polymerase III' consists of two DNA polymerase III and two t subunts. This data supports the idea that t may allow the polymerase III enzyme to dimerize providing a core upon which other polymerase auxiliary subunits assemble.
McHenry, C. (1981) J. Biol Chem. 257. 2657-2663.

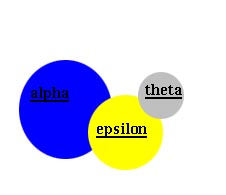
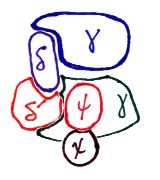
The above pictures are a schematic representation of DNA polymerase III core.
At left, are the core, tau, gamma complex, and beta sliding clamp.
Center, are the alpha, epsilon and theta subunits.
At right, the complex of gamma (2), delta, delta prime, psi, and chi subunits.
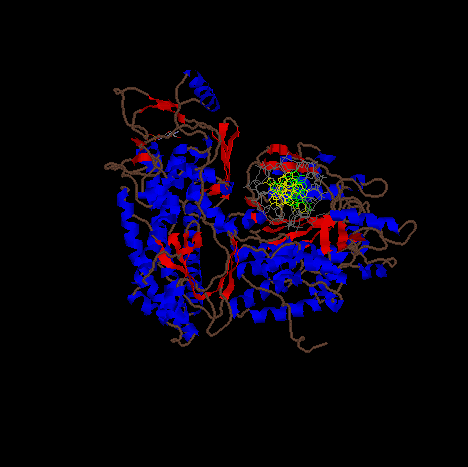
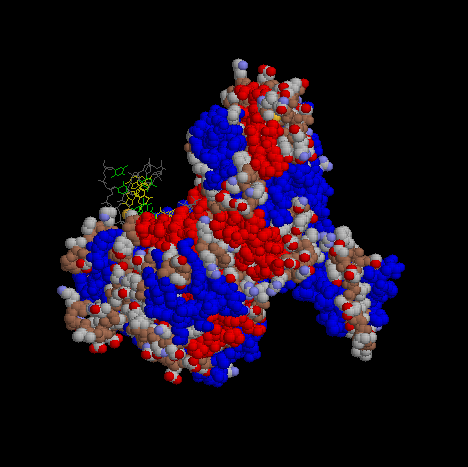
Image of DNA pol III complexed with a strand of DNA (yellow, green, and gray).
The image is a cartoon representation with helical segments of the protein are shown in blue and beta sheet regions are shown in red.
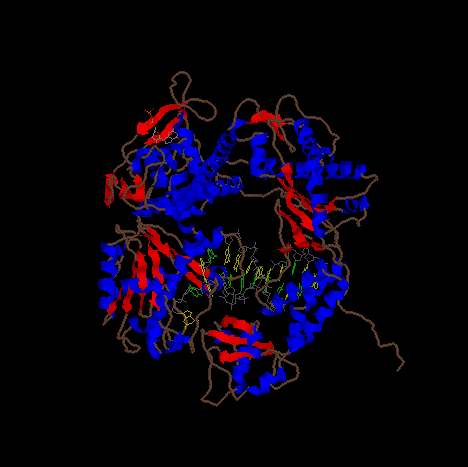
Image of DNA pol III complexed with a strand of DNA rotated to show more of the structure.
The image is a cartoon representation with helical segments of the protein are shown in blue and beta sheet regions are shown in red.
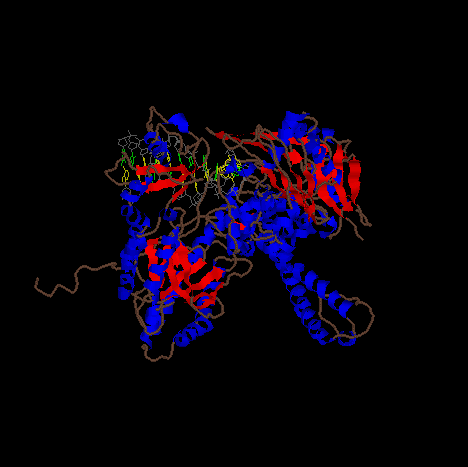
Same as above from a third angle.
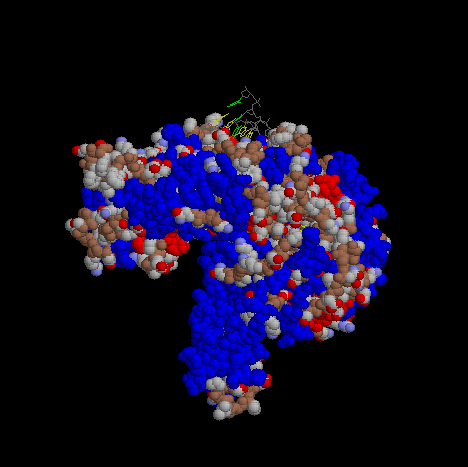
In order to show more of the three dimensional features of DNA pol III, a Spacefilling representation was used here and below at different angles.